Local Ancestry‐Informed Candidate Pathway Analysis of Warfarin Stable Dose in Latino Populations
So excited to see this publication finally come out. This was my first project I worked on with the Karnes Lab and I really think it became a diamond from the rough. I want to highlight two special things about this manuscript: the novel Manhattan plots and publication in a DEI journal.
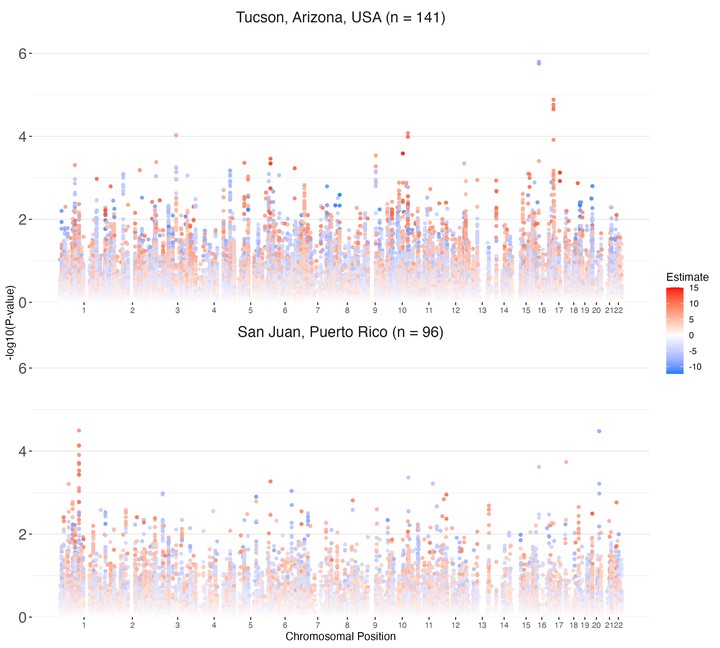
This project features the novel Manhattan plots that I designed to update the classic visualization to include additional biostatistical information. For over 10 years, Manhattan plots have been used to display the statistical results of genome-wide association studies (GWAS). These traditional plots, however, rely solely on chromosomal location and p-value of a statistical test, as displayed in the wikipedia example Manahattan Plot. Biostatistically and clinically, a p-value is of little importance without an effect size. I updated these classic plots to display effect sizes through color at each loci being visualized. I argue that no information is lost, while valuable statistical insight is gained, because chromosomal location remains mapped on the x-axis though no longer mapped by color. I am excited to report I have seen that code be utilized in other research outside of the Karnes Lab already (see Figure 3 here)!!
This work will be published in print in a special Diversity, Equity & Inclusion (DEI) issue of CP&T in March of this year which absolutely tickles my social justice soul. I started this research project quite uneasy about performing a genetic study in a Latine population because I couldn’t wrap my head around how being Latine could be genetic in any way. Though ultimately unable to sway the lab away from race/ethnicity-based research methods, in this manuscript I was able to solidify through data some important points to ultimately recommend against the clinical use of race/ethicnitiy in medicine. First, the populations of people who are categorized as Latine look vastly genetically different across the globe, thus, the categorization “Latine” will not be meaningful for clinical prediction. Second, the crux of the GWAS is that the method is hypothesis generating: every association uncovered through GWAS requires further validation. My argument is that smaller, genetically-diverse cohorts can be utilized without fear of population stratification because false positives are commonplace in GWAS. We find here the same associations published by much larger warfarin studies by using more advanced statistical procedures in lieu of recruiting larger populations.
I excitedly offered to give a talk on this research project at the Pharmacogenetics Global Research Network early career research in progress meeting in February so I can talk directly to the research community about this important work.