Local Ancestry‐Informed Candidate Pathway Analysis of Warfarin Stable Dose in Latino Populations
Abstract
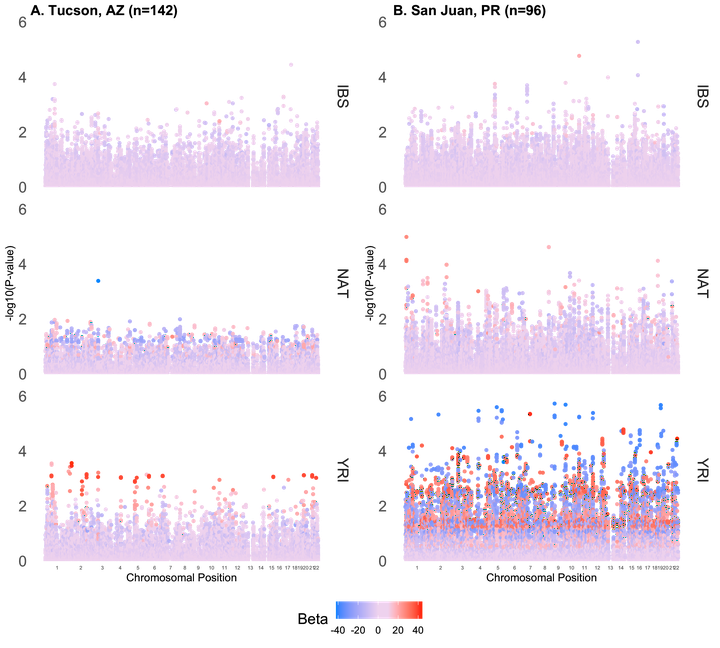
Accuracy of warfarin dose prediction algorithms may be improved by including data from diverse populations in genetic studies of dose variability. Here, we surveyed single nucleotide polymorphisms in vitamin K-related genetic pathways for association with warfarin dose requirements in two admixed Latino populations in standard-principal component adjusted and contemporary-local ancestry adjusted regression models. A total of five variants from vitamin K-related genes/pathways were associated with warfarin dose in both cohorts (P < 0.0125) in standard models. Local ancestry-adjusted analysis unveiled 35 associated variants with absolute effects ranging from β = 9.04 ( ±2.23) to 39.18 ( ±10.89) per ancestral allele in the discovery cohort and β = 6.47 (± 2.02) to 17.82 (± 6.83) in the replication cohort. Importantly, we demonstrate the technical validity of the Tractor model in cohorts with admixed ancestry from three founder populations and bring attention to the technical hurdles obstructing the inclusion of diverse, especially admixed, populations in pharmacogenomic research.
My Contribution
This is a very fun publication for me because I championed nearly every aspect of this clinical research project’s life cycle. I managed Institutional Review Board documentation, recruited and consented study participants, collected study data from Electronic Health Records, extracted DNA from bio-specimen, prepared array data for analysis with quality control and imputation, directed and performed bioinformatic pipelines and analyzed statistical results for this research project. Apart from proposing this clinical research and obtaining funding, I led this project to it’s successful publication in Clinical Pharmacology & Therapeutics.